Wastewater Based Epidemiology (WBE) Lab & Testing
Background
As a result of a generous donation from Jesus Tovar of T&P Farms and funding from Yuma County, The Yuma Center of Excellence for Desert Agriculture (YCEDA) established a biosafety level 2 laboratory at the Yuma Agricultural Center for wastewater-based epidemiology (WBE) in 2020 and launched weekly wastewater testing in Yuma County, in collaboration with Dr. Ian Pepper at the UArizona WEST Center, to help detect COVID-19 infection, mitigate the spread of the virus, and advert outbreaks in Yuma communities. Additional funding from Yuma County and Arizona Department of Health Services supported pilot projects to expand the weekly testing and assess WBE as an early-warning tool for identifying high or increasing COVID-19 disease prevalence throughout Yuma County. Funding from the State of Arizona, Office of the Governor enhanced the lab capabilities enabling YCEDA to incorporate near real-time SARS-CoV-2 variant testing through genome sequencing into ongoing efforts improving monitoring efforts and allowing for identification of new variants.
Dr. Bradley Schmitz, a national expert in organizing and executing WBE programs, aids in developing protocols for wastewater testing for SARS-CoV-2 quantification, data modeling and interpretation. A county-wide steering committee was established to guide and implement public health response and communication.
Wastewater-Based Epidemiology
Wastewater-based epidemiology is the public health tool of the future. An early warning tool that harnesses public waste, wastewater-based surveillance prevents infections and as a result, saves money and, more importantly, lives. We are building an infrastructure in Arizona that improves public health and the viability of businesses within Arizona and beyond.
Wastewater-Based Epidemiology Research
First proposed to monitor communities for illicit pharmaceuticals with “non-invasive” methods, wastewater-based epidemiology became mainstream globally during the COVID-19 pandemic. Researchers and public health officials expanded the tool’s utility to monitor for the presence of other consequential infectious diseases, such as polio and influenza. The potential applications in public health are limitless. Indeed, wastewater-based epidemiology can track disease trends, enumerate affected individuals, and inform a nimble public health policy. Further, we have barely scratched the surface of wastewater applications to promote One Health efforts, including those focusing on animal health.
Research Highlights: Publications and Links
Affiliates at the Yuma Center for Excellence in Desert Agriculture have published in high-impact scientific journals. Examples include.
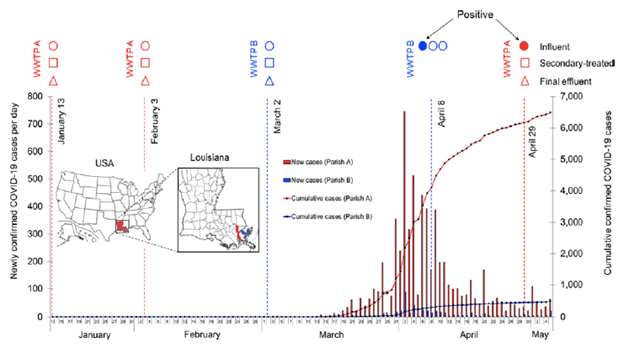
1. Sherchan SP, Shahin S, Ward LM, Tandukar S, Aw TG, Schmitz B, et al. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA - PubMed Sci Total Environ. 2020 Nov;743:140621
We investigated the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in wastewater samples in southern Louisiana, USA. To our knowledge, this was the first study reporting the detection of SARS-CoV-2 RNA in wastewater in North America. However, concentration methods and RT-qPCR assays need to be refined and validated to increase the sensitivity of SARS-CoV-2 RNA detection in wastewater.
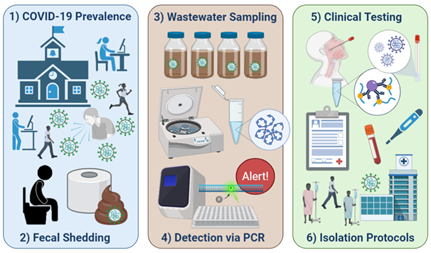
2. Betancourt WQ, Schmitz BW, Innes GK, Prasek SM, Pogreba Brown KM, Stark ER, et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention - PubMed Sci Total Environ. 2021;779:146408. .
The University of Arizona (UArizona) utilized WBE paired with clinical testing as a surveillance tool to monitor the UArizona community for SARS-CoV-2 in near real-time, as students re-entered campus in the fall of 2020. Positive detection of virus RNA in wastewater lead to selected clinical testing, identification, and isolation of three infected individuals (one symptomatic and two asymptomatic) that averted potential disease transmission. This case study demonstrated the value of WBE as a tool to efficiently utilize resources for COVID-19 prevention and response.
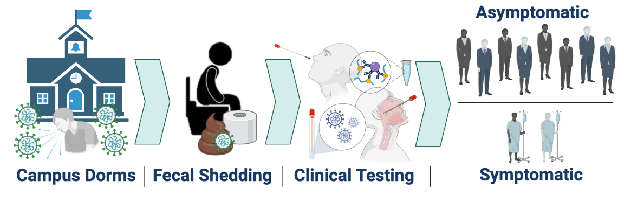
3. Schmitz BW, Innes GK, Prasek SM, Betancourt WQ, Stark ER, Foster AR, et al. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology - PubMed Sci Total Environ. 2021 Aug 20;801:149794.
Wastewater-based epidemiology was utilized to monitor SARS-CoV-2 RNA in sewage collected from manholes specific to individual student dormitories at the University of Arizona in the fall semester of 2020, which led to successful identification and reduction of SARS-CoV-2 transmission events. Positive wastewater samples triggered clinical testing of residents within that dorm; thus, SARS-CoV-2 infected individuals were identified regardless of symptom expression. Nasal and nasopharyngeal swab samples processed via antigen and PCR tests indicated that 79.2% of SARS-CoV-2 infections were asymptomatic, and only 20.8% of positive cases reported COVID-19 symptoms at the time of testing. Clinical data were paired with corresponding wastewater virus concentrations, which enabled calculation of viral shedding rates in feces per infected person. Mean shedding rates averaged from positive wastewater samples across all dorms were 7.30 ± 0.67 log10 genome copies per gram of feces (gc/g-feces) based on the N1 gene.
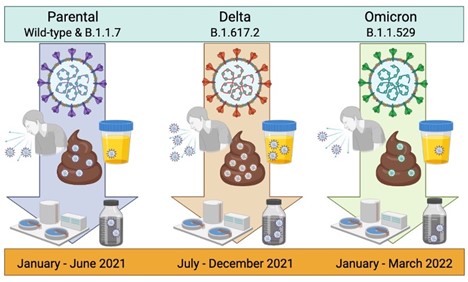
4. Prasek SM, Pepper IL, Innes GK, Slinski S, Betancourt WQ, Foster AR,…,Schmitz BW. Variant-specific SARS-CoV-2 shedding rates in wastewater - PubMed Sci Total Environ. 2023;857:159165.
Mean SARS-CoV-2 waste shedding rates were found to increase with the predominance of the Delta variant and subsequently decrease with Omicron infections. The Delta stage had the highest mean shedding rates and was associated with the most severe disease symptoms reported in other clinical studies, while Omicron, exhibiting reduced symptoms, had the lowest mean shedding rates. Additionally, shedding rates were most consistent across communities during the Omicron stage. This is the first paper to identify waste shedding rates specific to the Omicron variant and fills a knowledge gap critical to disease prevalence modeling.
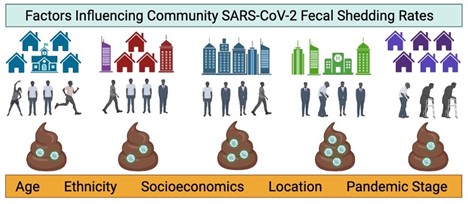
5. Prasek SM, Pepper IL, Innes GK, Slinski S, Ruedas M, Sanchez A,…, Schmitz BW. Population level SARS-CoV-2 fecal shedding rates determined via wastewater-based epidemiology - PubMed Sci Total Environ. 2022 Sep;838(Pt 4):156535.
Wastewater from six municipalities in Arizona and Florida with distinct demographics were monitored for SARS-CoV-2 RNA between September 2020 and December 2021. Average SARS-CoV-2 RNA fecal shedding rates were unique to each community and influenced by population demographics. Age, ethnicity, and socio-economic factors may have influenced shedding rates. Findings in this study suggest that community-specific shedding rates may be appropriate in model development relating wastewater virus concentrations to clinical case counts.
6. Casey JA, Tartof SY, Davis MF, Nachman KE, Price L, Liu C, Yu K, Gupta V, Innes GK, Tseng HF, Do V, Pressman AR, and Rudolph KE. Impact of a Statewide Livestock Antibiotic Use Policy on Resistance in Human Urine Escherichia coli Isolates: A Synthetic Control Analysis
In the United States, antimicrobial-resistant (AMR) pathogens cause nearly 3million3million infections and 35,000 deaths annually. Despite modest declines, sales of antimicrobials for food-animal production represent approximately 65% of antibiotics sold in the United States. Antimicrobial-resistant pathogens may spread from industrial food animal production sites to people through multiple routes, including foodborne, occupational, and environmental exposures. Animals can harbor bacteria from the farm to the processing facility where they can contaminate the retail meat supply, where improperly handled or undercooked meat can lead to human infection.
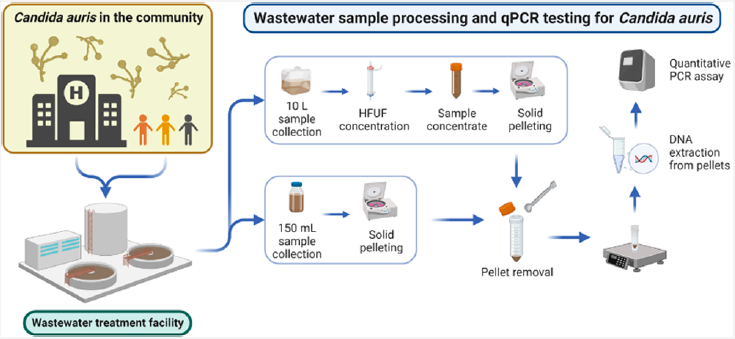
7. Barber C, Crank K, Papp K, Innes GK, Schmitz BW, Chavez J, Rossi A, and Gerrity D. Community-Scale Wastewater Surveillance of Candida auris during an Ongoing Outbreak in Southern Nevada
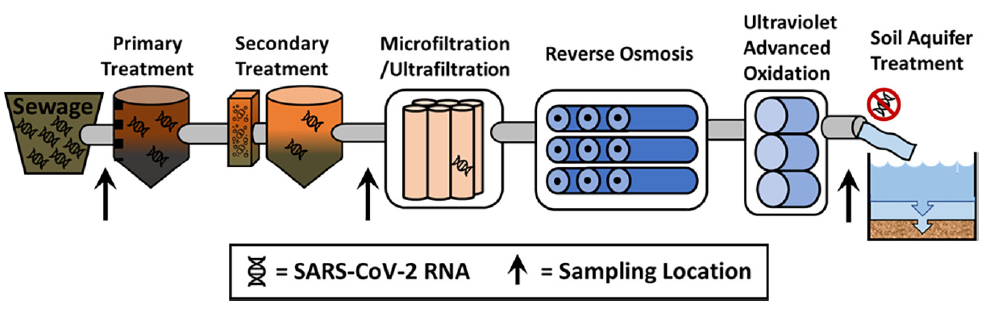
8. Polanco J, Schmitz BW, Choi S, Safarik J, Prasek S, and Plumlee MH. SARS-CoV-2 genetic material is removed during municipal wastewater treatment and is undetectable after advanced treatment
One Health
-
Jesus Tovar (T&P Farms)
-
Yuma County
-
Arizona Department of Health Services
-
State of Arizona
-
DatePac LLC
-
Translational Genomics Research Institute
Related Videos
 Jan 13, 2023
Jan 13, 20232023 YCEDA Research Summary
 May 17, 2022
May 17, 2022Paul Brierley: Wastewater Surveillance Informs Public Health Actions to Prevent Disease in Yuma, AZ
 May 04, 2022
May 04, 2022Poop Doesn't Lie: How Wastewater Testing Became a Global Public Health Tool